Bayesian estimation techniques were added to Mplus version 7 some time ago. Of course I was interested if MPlus and JAGS come to comparable results when let loose on data. 😉
In particular I was interested in the crossclassified module of MPlus, so I wrote some code to simulate data with R and subsequently recovered the parameters with JAGS and MPlus. Recently I found out how JAGS could also be used as a data simulator for simulation studies, but this is a different topic.
So graphically, the model I used for simulation looks like this:
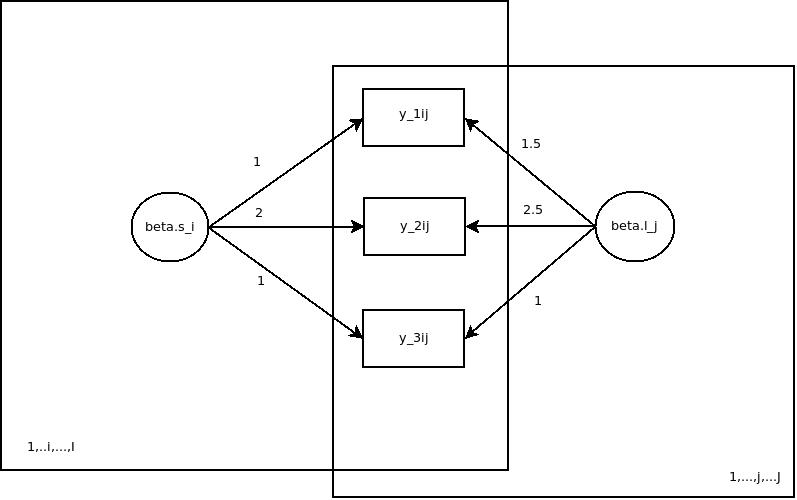
I have omitted the level-1 residual variances and the variances of the latent distributions in the figure for simplicity reasons. The full model specifications can be gleaned from the source code attached to this post.
To make a long story shot: both approaches, JAGs and MPlus, pleasently come to very comparable results with regard to the parameters. At the moment I am too busy with other things to harp on this more, but I thought it is useful to put the code on the blog in case someone is interested to work on a similar topic in more detail. Thanks to Mr Asparouhov for clarifying some questions with regard to MPlus.
The files:
test.R: code to simulate data from the model, recover the parameters with JAGs, also a bit of convergence diagnostics
test.txt: JAGs model file
cc.inp: MPlus input file
cc.dat: Simulated data. Will be overwritten once the simulation part of test.R is executed.
Get the files here. The code is not very polished but should give an idea how to specify the model in MPlus and JAGs.
